Overview
The measure package provides a comprehensive suite of
functions for analytical method validation. These functions are designed
to be compatible with regulatory frameworks including: - ICH
Q2(R2): Validation of Analytical Procedures - ISO/IEC
17025: General requirements for testing and calibration
laboratories - USP <1225>: Validation of
Compendial Procedures - ICH M10: Bioanalytical Method
Validation (for applicable workflows)
This vignette demonstrates key validation workflows including calibration, precision, accuracy, uncertainty, and quality control.
Calibration Curves
Fitting Calibration Curves
The measure_calibration_fit() function fits weighted or
unweighted calibration curves with comprehensive diagnostics.
# Create calibration data
set.seed(42)
cal_data <- data.frame(
nominal_conc = c(1, 5, 10, 25, 50, 100, 250, 500),
response = c(1, 5, 10, 25, 50, 100, 250, 500) * 1.05 +
rnorm(8, sd = c(0.1, 0.3, 0.5, 1, 2, 4, 10, 20))
)
# Fit with 1/x^2 weighting (common for bioanalytical methods)
cal <- measure_calibration_fit(
cal_data,
response ~ nominal_conc,
weights = "1/x2"
)
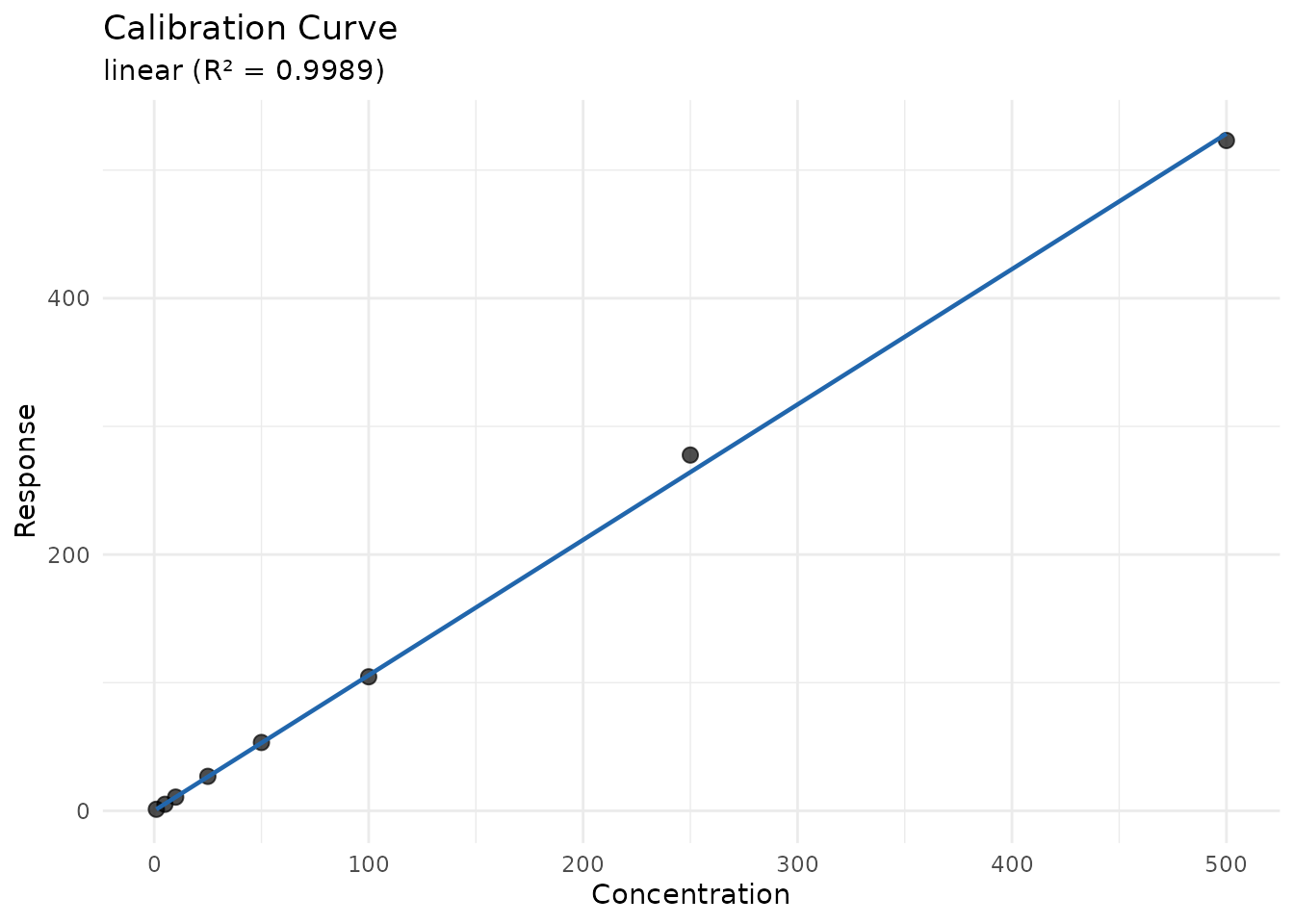
print(cal)
#> <measure_calibration>
#> Model: linear
#> Weighting: 1/x2
#> Formula: response ~ nominal_conc
#> N points: 8
#> R²: 0.9989Predicting Unknown Concentrations
unknowns <- data.frame(
sample_id = c("Sample_1", "Sample_2", "Sample_3"),
response = c(52.3, 125.8, 280.5)
)
predictions <- measure_calibration_predict(
cal,
newdata = unknowns,
interval = "confidence"
)
cbind(unknowns, predictions)
#> sample_id response .pred_conc .pred_lower .pred_upper
#> 1 Sample_1 52.3 49.3896 49.30708 49.47213
#> 2 Sample_2 125.8 118.9571 118.87457 119.03962
#> 3 Sample_3 280.5 265.3801 265.29758 265.46263Calibration Verification
Verify that the calibration remains valid using QC samples:
qc_data <- data.frame(
sample_id = c("QC_Low", "QC_Mid", "QC_High"),
nominal_conc = c(3, 75, 400),
response = c(3.1, 77.5, 395.2)
)
verification <- measure_calibration_verify(cal, qc_data)
print(verification)
#>
#> ── Calibration Verification ────────────────────────────────────────────────────
#> ✔ Overall: PASS (3/3 samples within 15%)
#>
#> ── Sample Results ──
#>
#> # A tibble: 3 × 8
#> sample_id nominal_conc response predicted_conc accuracy_pct deviation_pct
#> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 QC_Low 3 3.1 2.82 94.1 -5.93
#> 2 QC_Mid 75 77.5 73.2 97.7 -2.34
#> 3 QC_High 400 395. 374. 93.5 -6.51
#> # ℹ 2 more variables: acceptance_limit <dbl>, pass <lgl>Limits of Detection and Quantitation
Multiple Methods
measure supports multiple approaches for calculating
LOD/LOQ:
# Blank-based approach (3σ/10σ)
blank_data <- data.frame(
sample_type = rep("blank", 10),
response = rnorm(10, mean = 0.5, sd = 0.08)
)
lod_result <- measure_lod(
blank_data,
"response",
method = "blank_sd",
calibration = cal
)
print(lod_result)
#> <measure_lod>
#> Value: 0.8726
#> Method: blank_sd
#> k: 3
#> Uncertainty: 0.1142
#> Parameters:
#> blank_mean: 0.5115
#> blank_sd: 0.1203
#> n_blanks: 10
# Or calculate both together
lod_loq <- measure_lod_loq(
blank_data,
"response",
method = "blank_sd",
calibration = cal
)
tidy(lod_loq)
#> # A tibble: 2 × 5
#> limit_type value method k uncertainty
#> <chr> <dbl> <chr> <dbl> <dbl>
#> 1 LOD 0.873 blank_sd 3 0.114
#> 2 LOQ 1.72 blank_sd 10 0.381Precision Studies
Repeatability (Within-Run Precision)
# Data from replicate measurements
repeat_data <- data.frame(
sample_id = rep(c("Low", "Mid", "High"), each = 6),
concentration = c(
rnorm(6, 10, 0.5),
rnorm(6, 100, 4),
rnorm(6, 500, 18)
)
)
repeatability <- measure_repeatability(
repeat_data,
"concentration",
group_col = "sample_id"
)
print(repeatability)
#> measure_precision: repeatability
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> Group: Low
#> n = 6
#> Mean = 9.82
#> SD = 0.7645
#> CV = 7.8 %
#> 95% CI: [9.017, 10.62]
#>
#> Group: Mid
#> n = 6
#> Mean = 99.51
#> SD = 4.893
#> CV = 4.9 %
#> 95% CI: [94.37, 104.6]
#>
#> Group: High
#> n = 6
#> Mean = 501.1
#> SD = 18.58
#> CV = 3.7 %
#> 95% CI: [481.6, 520.6]Intermediate Precision
# Data from multiple days
ip_data <- data.frame(
day = rep(1:3, each = 6),
analyst = rep(c("A", "A", "A", "B", "B", "B"), 3),
concentration = 100 +
rep(c(-2, 0, 2), each = 6) + # Day effect
rep(c(-1, 1), 9) + # Analyst effect
rnorm(18, sd = 3) # Residual
)
ip_result <- measure_intermediate_precision(
ip_data,
"concentration",
factors = c("day", "analyst")
)
print(ip_result)
#> measure_precision: intermediate
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> Variance Components:
#> day: 15.78 (57%)
#> analyst: 3.82 (14%)
#> Residual: 8.135 (29%)
#>
#> CV by component:
#> day: 4%
#> analyst: 2%
#> Residual: 2.9%Gage R&R Analysis
For measurement system analysis:
# Gage R&R data
grr_data <- data.frame(
part = rep(1:5, each = 6),
operator = rep(rep(c("Op1", "Op2"), each = 3), 5),
measurement = c(
# Part 1
10.1, 10.2, 10.0, 10.3, 10.1, 10.2,
# Part 2
20.2, 20.1, 20.3, 20.0, 20.2, 20.1,
# Part 3
15.1, 15.0, 15.2, 15.3, 15.1, 15.0,
# Part 4
25.0, 25.1, 24.9, 25.2, 25.0, 25.1,
# Part 5
30.1, 30.2, 30.0, 30.1, 30.0, 30.2
)
)
grr_result <- measure_gage_rr(
grr_data,
"measurement",
part_col = "part",
operator_col = "operator"
)
print(grr_result)
#> measure_gage_rr: Measurement System Analysis
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> Study design:
#> Parts: 5
#> Operators: 2
#> Replicates: 3
#>
#> Variance Components:
#> Repeatability: 0.01133 (0.02% contribution)
#> Reproducibility: 0 (0% contribution)
#> Total R&R: 0.01133 (0.02% contribution)
#> Part-to-Part: 62.08 (100% contribution)
#>
#> % Study Variation:
#> Repeatability: 1%
#> Reproducibility: 0%
#> Total R&R: 1%
#> Part-to-Part: 100%
#>
#> Number of Distinct Categories (ndc): 104
#>
#> Assessment:
#> Measurement system is ACCEPTABLE (%R&R < 10%)Accuracy Assessment
Bias and Recovery
accuracy_data <- data.frame(
level = rep(c("Low", "Mid", "High"), each = 5),
measured = c(
rnorm(5, 10.2, 0.3), # Low level, slight positive bias
rnorm(5, 100, 2.5), # Mid level, no bias
rnorm(5, 498, 8) # High level, slight negative bias
),
reference = rep(c(10, 100, 500), each = 5)
)
accuracy <- measure_accuracy(
accuracy_data,
"measured",
"reference",
group_col = "level"
)
print(accuracy)
#> measure_accuracy
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> Group: Low
#> n = 5
#> Mean measured = 10.09
#> Mean reference = 10
#> Bias = 0.08854 ( 0.89 %)
#> Recovery = 101 %
#> Recovery 95% CI: [95%, 106%]
#>
#> Group: Mid
#> n = 5
#> Mean measured = 101
#> Mean reference = 100
#> Bias = 1.042 ( 1 %)
#> Recovery = 101 %
#> Recovery 95% CI: [99%, 103%]
#>
#> Group: High
#> n = 5
#> Mean measured = 502.6
#> Mean reference = 500
#> Bias = 2.593 ( 0.52 %)
#> Recovery = 101 %
#> Recovery 95% CI: [99%, 102%]Linearity Assessment
linearity_data <- data.frame(
concentration = rep(c(10, 25, 50, 75, 100), each = 3),
response = rep(c(10, 25, 50, 75, 100), each = 3) * 1.02 +
rnorm(15, sd = 1.5)
)
linearity <- measure_linearity(
linearity_data,
"concentration",
"response"
)
print(linearity)
#> measure_linearity
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> Data:
#> n = 15 ( 5 levels )
#> Range: 10 - 100
#>
#> Regression:
#> Slope = 1.023
#> 95% CI: [1.003, 1.044]
#> Intercept = -0.1535
#> 95% CI: [-1.436, 1.129]
#>
#> Fit Quality:
#> R-squared = 0.99884
#> Adj. R-squared = 0.99875
#> Residual SD = 1.222
#> Residual CV = 2.3 %
#>
#> Lack-of-Fit Test:
#> F = 0.649
#> p-value = 0.6015
#> Result: Not significant (linearity acceptable)
# Plot with fit line
autoplot(linearity, type = "fit")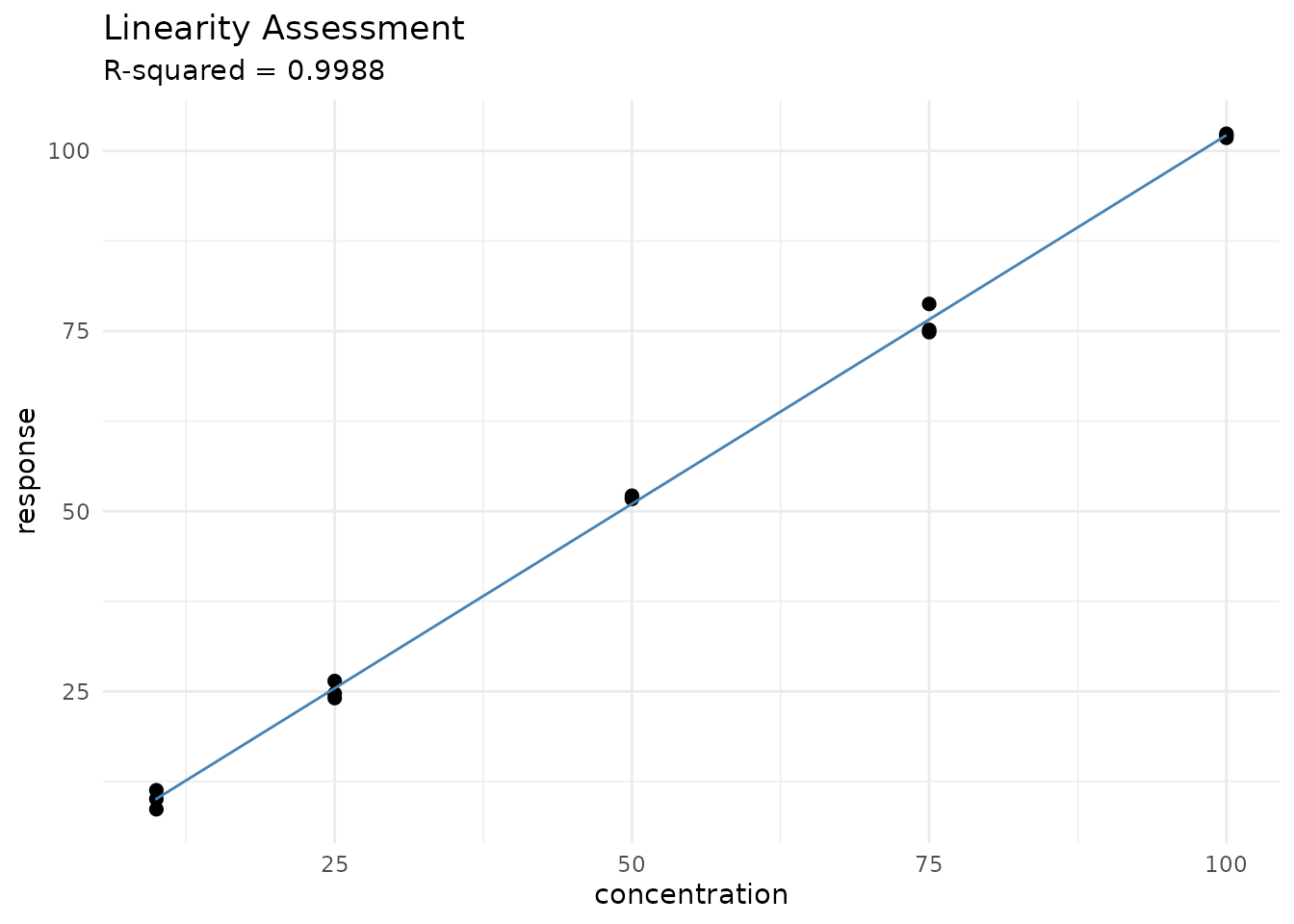
Uncertainty Budgets
ISO GUM Uncertainty
Create uncertainty budgets following the GUM (Guide to the Expression of Uncertainty in Measurement):
# Define uncertainty components
components <- list(
uncertainty_component(
name = "Repeatability",
type = "A",
value = 0.5,
df = 9
),
uncertainty_component(
name = "Calibration",
type = "B",
value = 0.3,
distribution = "normal"
),
uncertainty_component(
name = "Reference Standard",
type = "B",
value = 0.1,
distribution = "rectangular"
),
uncertainty_component(
name = "Temperature",
type = "B",
value = 0.2,
sensitivity = 0.5 # Sensitivity coefficient
)
)
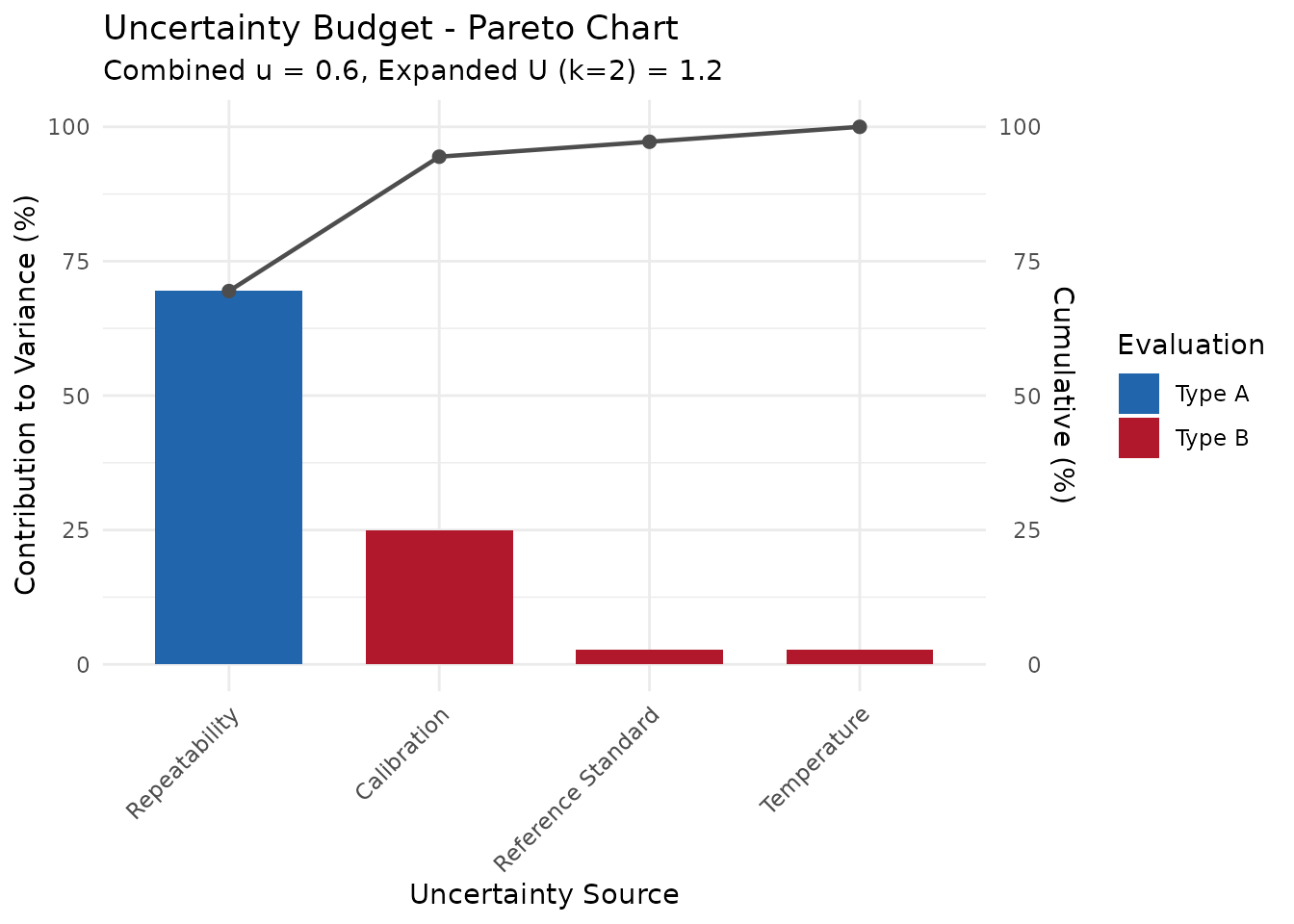
budget <- measure_uncertainty_budget(.list = components)
print(budget)
#> <measure_uncertainty_budget>
#> Components: 4 (1 Type A, 3 Type B)
#> Combined u: 0.6
#> Effective df: 19
#> Coverage k: 2
#> Expanded U: 1.2Control Charts
Setting Up Control Limits
# Historical QC data
qc_history <- data.frame(
run_order = 1:30,
qc_value = rnorm(30, mean = 100, sd = 2)
)
limits <- measure_control_limits(qc_history, "qc_value")
print(limits)
#> measure_control_limits: shewhart chart
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> n = 30
#> Center = 100.1
#> Sigma = 1.737
#> UCL (+3s) = 105.3
#> UWL (+2s) = 103.6
#> LWL (-2s) = 96.61
#> LCL (-3s) = 94.88Monitoring with Westgard Rules
# New run data including potential out-of-control point
new_run <- data.frame(
run_order = 1:20,
qc_value = c(rnorm(19, 100, 2), 108) # Last point is high
)
chart <- measure_control_chart(
new_run,
"qc_value",
"run_order",
limits = limits,
rules = c("1_3s", "2_2s", "R_4s", "10x")
)
print(chart)
#> measure_control_chart
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> Observations: 20
#> Rules applied: 1_3s, 2_2s, R_4s, 10x
#> Violations detected: 4
#>
#> Status: OUT OF CONTROL
#>
#> Violation summary:
#> # A tibble: 4 × 3
#> run_order qc_value violation
#> <int> <dbl> <chr>
#> 1 4 105. 1:3s R:4s
#> 2 5 97.3 R:4s
#> 3 19 100. R:4s
#> 4 20 108 1:3s R:4s
autoplot(chart)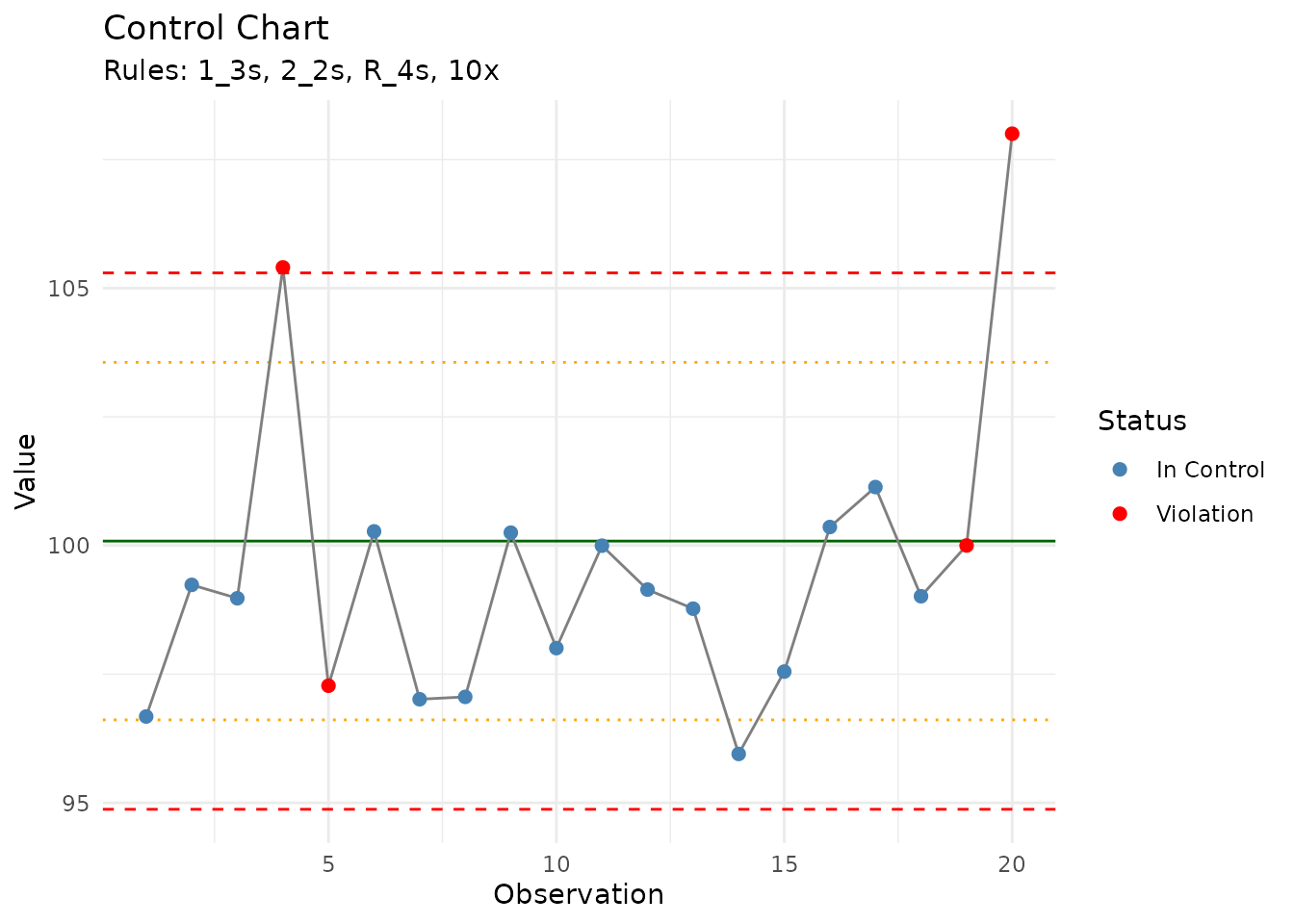
Acceptance Criteria
Defining Criteria
# Create custom criteria
my_criteria <- measure_criteria(
criterion("cv", "<=", 15, description = "Precision CV"),
criterion("bias_pct", "between", c(-10, 10), description = "Bias"),
criterion("recovery", "between", c(85, 115), description = "Recovery %")
)
print(my_criteria)
#> <measure_criteria> with 3 criteria
#> • Precision CV
#> • Bias
#> • Recovery %Using Preset Criteria
# ICH Q2 presets
ich_criteria <- criteria_ich_q2()
print(ich_criteria)
#> <measure_criteria> with 4 criteria
#> • Repeatability RSD <= 2%
#> • Intermediate precision RSD <= 5%
#> • Recovery 98-102%
#> • R² >= 0.999
# Bioanalytical presets
bio_criteria <- criteria_bioanalytical()
print(bio_criteria)
#> <measure_criteria> with 5 criteria
#> • QC CV <= 15%
#> • Calibration CV <= 20%
#> • R² >= 0.99
#> • Recovery 80-120%
#> • Bias within +/-15%Assessing Results
# Sample results to assess (single summary values per criterion)
# For example, from a method validation summary
results <- list(
cv = 5.2, # Overall precision CV
bias_pct = 1.3, # Overall bias
recovery = 101.3 # Mean recovery
)
assessment <- measure_assess(results, my_criteria)
print(assessment)
#> <measure_assessment> [PASS]
#> 3 passed, 0 failed
#>
#> ✓ cv: 5.2 (<= 15)
#> ✓ bias_pct: 1.3 (between [-10, 10])
#> ✓ recovery: 101.3 (between [85, 115])
# Check if all criteria passed
all_pass(assessment)
#> [1] TRUEMethod Comparison
When validating a new method, you often need to compare it against a
reference or existing method. The measure package provides
several approaches for method comparison studies.
Bland-Altman Analysis
Bland-Altman plots show the agreement between two methods by plotting differences against means:
# Paired measurements from two methods
comparison_data <- data.frame(
sample_id = 1:30,
method_A = rnorm(30, mean = 100, sd = 15),
method_B = rnorm(30, mean = 102, sd = 16)
)
ba <- measure_bland_altman(
comparison_data,
method1_col = "method_A",
method2_col = "method_B",
regression = "linear" # Test for proportional bias
)
print(ba)
#> measure_bland_altman
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> Bias Statistics:
#> n = 30
#> Mean bias = 4.083
#> SD of differences = 17.97
#> 95% CI for bias: [-2.625, 10.79]
#>
#> Limits of Agreement:
#> Lower LOA = -31.13 (95% CI: [ -42.75 , -19.51 ])
#> Upper LOA = 39.3 (95% CI: [ 27.68 , 50.92 ])
#> LOA Width = 70.43
#>
#> Proportional Bias Test:
#> Slope = 0.2046
#> p-value = 0.57
#> Result: No significant proportional bias
autoplot(ba)
#> `geom_smooth()` using formula = 'y ~ x'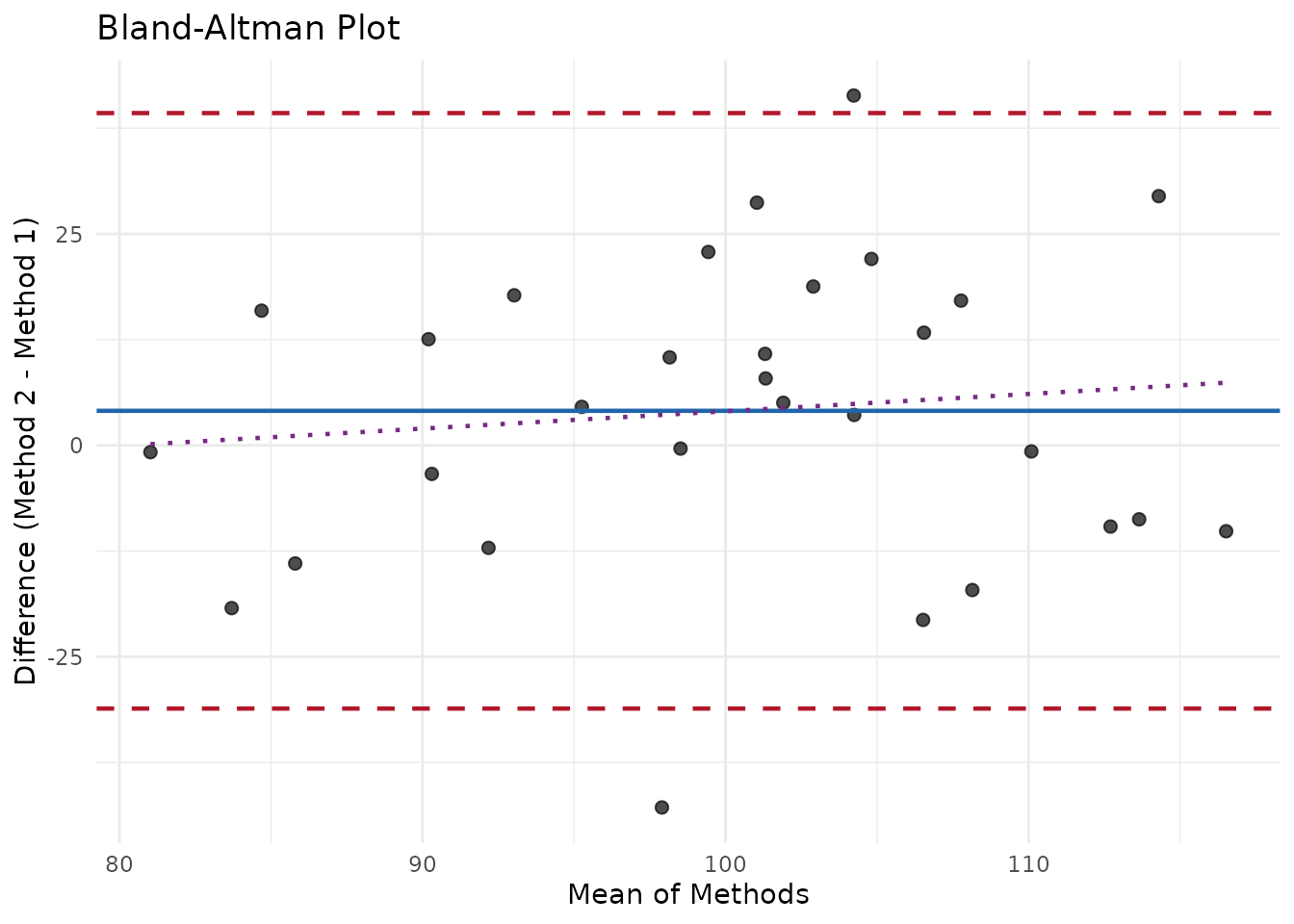
Regression Methods
For method comparison regression, use Deming or Passing-Bablok regression which account for error in both methods:
# Method comparison with known measurement error
deming_data <- data.frame(
reference = c(5, 10, 25, 50, 100, 200, 400),
test_method = c(5.2, 10.3, 25.8, 51.2, 101.5, 203.1, 408.2)
)
deming <- measure_deming_regression(
deming_data,
method1_col = "reference",
method2_col = "test_method",
bootstrap = TRUE,
bootstrap_n = 500
)
#> Using default error ratio of 1. Provide `error_ratio` or SDs for more accurate
#> results.
print(deming)
#> measure_deming_regression
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> Coefficients:
#> # A tibble: 2 × 4
#> term estimate ci_lower ci_upper
#> <chr> <dbl> <dbl> <dbl>
#> 1 intercept -0.00414 -0.325 0.437
#> 2 slope 1.02 1.01 1.02
#>
#> Statistics:
#> n = 7
#> Error ratio = 1
#> RMSE = 0.4087
#> R² = 1
#>
#> (Fitted using mcr package)
# Check if methods are equivalent
glance(deming)
#> # A tibble: 1 × 6
#> intercept slope intercept_ci_includes_0 slope_ci_includes_1 r_squared rmse
#> <dbl> <dbl> <lgl> <lgl> <dbl> <dbl>
#> 1 -0.00414 1.02 TRUE FALSE 1.000 0.409For Passing-Bablok regression (non-parametric), install the
mcr package:
# Requires: install.packages("mcr")
pb <- measure_passing_bablok(
deming_data,
method1_col = "reference",
method2_col = "test_method"
)
print(pb)Proficiency Testing
Evaluate laboratory performance in proficiency testing programs:
# PT results from multiple labs
pt_data <- data.frame(
lab_id = paste0("Lab_", 1:10),
measured = c(99.2, 100.5, 98.8, 101.2, 97.5, 100.1, 99.8, 102.3, 100.6, 94.0),
assigned = rep(100, 10),
uncertainty = c(1.5, 2.0, 1.8, 1.6, 2.2, 1.9, 1.7, 2.1, 1.5, 2.0)
)
# z-scores with known sigma
z_scores <- measure_proficiency_score(
pt_data,
measured_col = "measured",
reference_col = "assigned",
score_type = "z_score",
sigma = 2.5
)
print(z_scores)
#> measure_proficiency_score
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> Score Type: z_score
#> Sigma: 2.5
#>
#> Results (n = 10 ):
#> Satisfactory (|z| ≤ 2): 9 ( 90 %)
#> Questionable (2 < |z| ≤ 3): 1
#> Unsatisfactory (|z| > 3): 0
#>
#> Score Statistics:
#> Mean score: -0.24
#> SD score: 0.925
#> Max |score|: 2.4
autoplot(z_scores)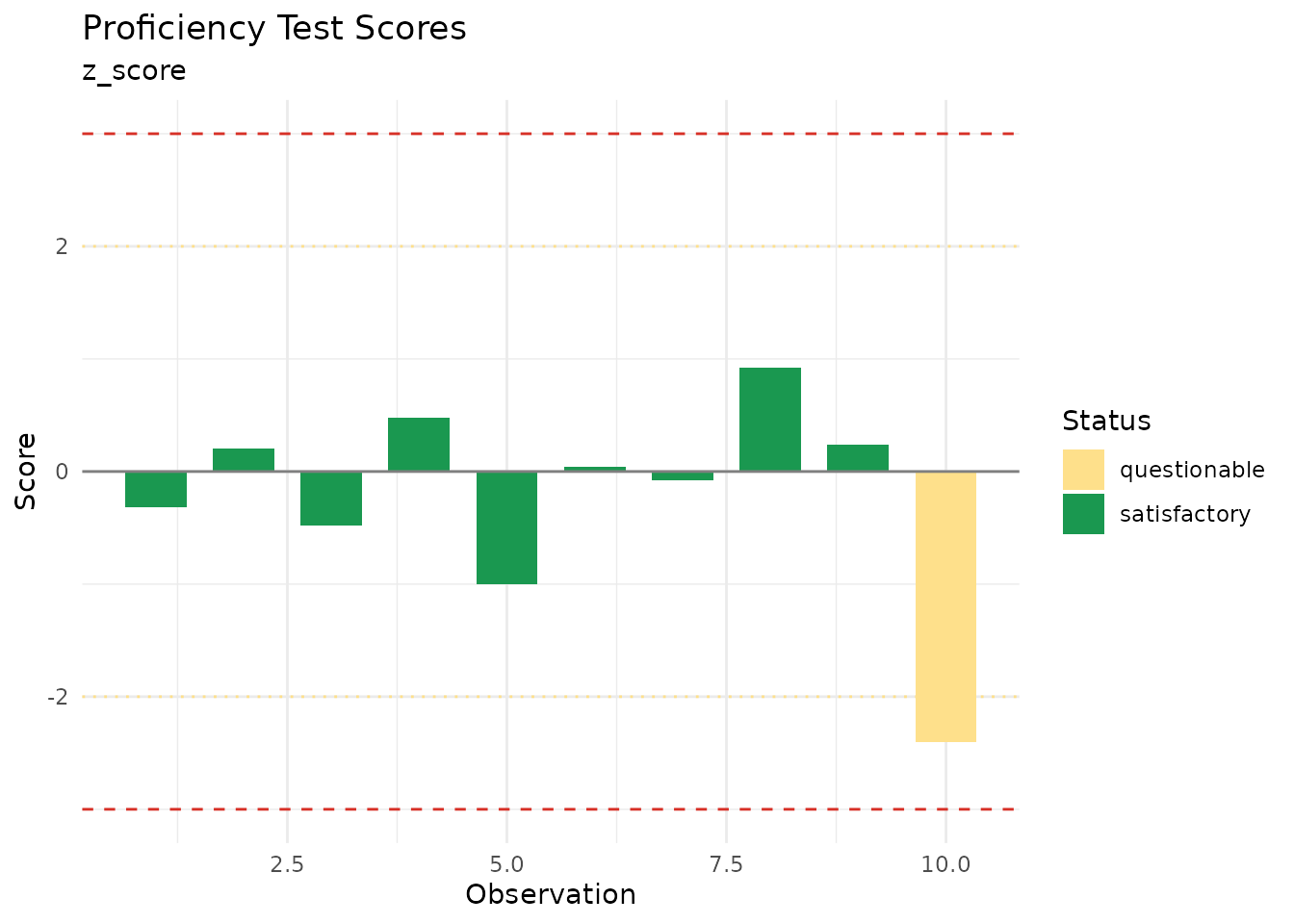
Matrix Effects
Matrix effects (ion suppression/enhancement) must be evaluated in LC-MS/MS and similar methods.
Evaluating Matrix Effects
# Post-extraction spike experiment
me_data <- data.frame(
sample_type = rep(c("matrix", "neat"), each = 6),
matrix_lot = rep(c("Lot1", "Lot2", "Lot3"), 4),
concentration = rep(c("low", "high"), each = 3, times = 2),
response = c(
# Matrix samples (some suppression)
9200, 9500, 8900, 47500, 48200, 46800,
# Neat samples
10000, 10000, 10000, 50000, 50000, 50000
)
)
me <- measure_matrix_effect(
me_data,
response_col = "response",
sample_type_col = "sample_type",
matrix_level = "matrix",
neat_level = "neat",
concentration_col = "concentration"
)
print(me)
#> measure_matrix_effect
#> ────────────────────────────────────────────────────────────────────────────────
#>
#> Overall Matrix Effect Summary:
#> Groups evaluated: 2
#> Mean ME: 94 %
#> SD ME: 2 %
#> CV ME: 2 %
#> Range: 92 - 95 %
#>
#> Effect Classification:
#> Ion suppression (ME < 100%): 2
#> Ion enhancement (ME > 100%): 0
#> Acceptable (80-120%): 2 / 2
autoplot(me, type = "bar")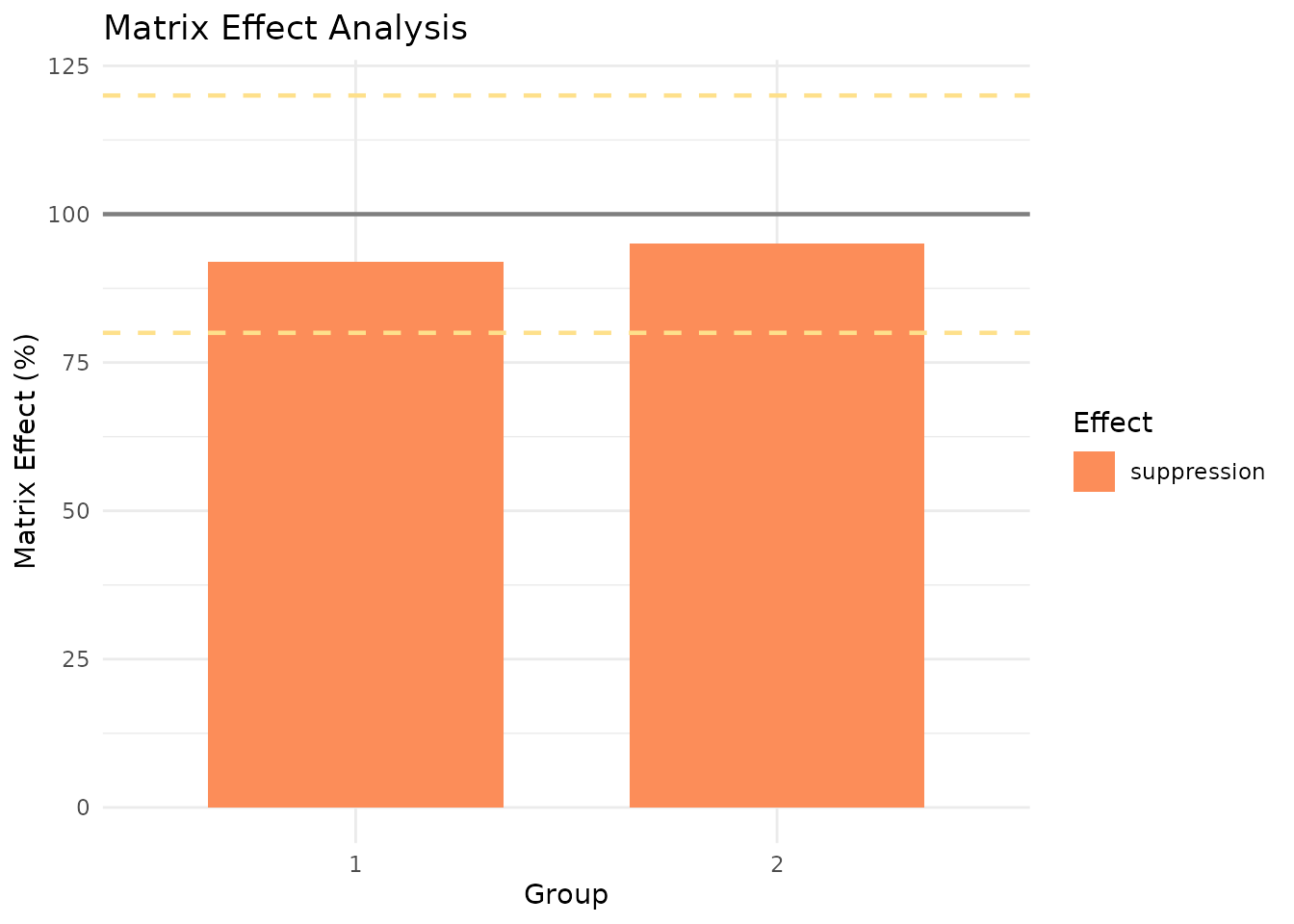
Standard Addition Correction
When matrix effects vary between samples, standard addition provides sample-specific correction:
library(recipes)
# Standard addition data
sa_data <- data.frame(
sample_id = rep(c("Sample1", "Sample2"), each = 4),
addition = rep(c(0, 10, 20, 30), 2),
response = c(
150, 250, 350, 450, # Sample 1
250, 350, 450, 550 # Sample 2
)
)
rec <- recipe(~ ., data = sa_data) |>
step_measure_standard_addition(
response,
addition_col = "addition",
sample_id_col = "sample_id"
) |>
prep()
# Original concentrations calculated via extrapolation
bake(rec, new_data = NULL)Sample Preparation QC
Recipe steps for quality control during sample preparation.
Dilution Factor Correction
Back-calculate concentrations for diluted samples:
library(recipes)
dilution_data <- data.frame(
sample_id = paste0("S", 1:5),
dilution_factor = c(1, 2, 5, 10, 1),
analyte = c(50, 45, 42, 48, 51) # Measured after dilution
)
rec <- recipe(~ ., data = dilution_data) |>
update_role(sample_id, new_role = "id") |>
step_measure_dilution_correct(
analyte,
dilution_col = "dilution_factor",
operation = "multiply"
) |>
prep()
# Back-calculated original concentrations
bake(rec, new_data = NULL)
#> # A tibble: 5 × 3
#> sample_id dilution_factor analyte
#> <chr> <dbl> <dbl>
#> 1 S1 1 50
#> 2 S2 2 90
#> 3 S3 5 210
#> 4 S4 10 480
#> 5 S5 1 51Surrogate Recovery
Monitor extraction efficiency with surrogate standards:
qc_data <- data.frame(
sample_id = paste0("QC", 1:6),
surrogate = c(95, 105, 88, 112, 75, 132) # Expected = 100
)
rec <- recipe(~ ., data = qc_data) |>
update_role(sample_id, new_role = "id") |>
step_measure_surrogate_recovery(
surrogate,
expected_value = 100,
action = "flag",
min_recovery = 80,
max_recovery = 120
) |>
prep()
# Flag samples outside recovery limits
bake(rec, new_data = NULL)
#> # A tibble: 6 × 3
#> sample_id surrogate .surrogate_pass
#> <chr> <dbl> <lgl>
#> 1 QC1 95 TRUE
#> 2 QC2 105 TRUE
#> 3 QC3 88 TRUE
#> 4 QC4 112 TRUE
#> 5 QC5 75 FALSE
#> 6 QC6 132 FALSEDrift Correction
Detecting Drift
# Data with drift
drift_data <- data.frame(
sample_type = rep("qc", 20),
run_order = 1:20,
feature1 = 100 + (1:20) * 0.8 + rnorm(20, sd = 2), # Has drift
feature2 = 100 + rnorm(20, sd = 2) # No drift
)
drift_result <- measure_detect_drift(
drift_data,
features = c("feature1", "feature2"),
qc_type = "qc"
)
print(drift_result)
#> # A tibble: 2 × 5
#> feature slope slope_pvalue percent_change significant
#> <chr> <dbl> <dbl> <dbl> <lgl>
#> 1 feature1 0.630 0.0000148 11.1 TRUE
#> 2 feature2 -0.0351 0.634 -0.662 FALSEValidation Reports
Once you’ve completed your validation studies, you can compile all
results into a reproducible validation report using
measure_validation_report(). The package provides templates
following regulatory frameworks like ICH Q2(R2) and USP <1225>.
### Creating a Validation Report
# Gather validation results (using objects from above)
report <- measure_validation_report(
# Metadata
title = "HPLC-UV Method Validation Report",
method_name = "Compound X Assay",
method_description = "Reversed-phase HPLC with UV detection at 254 nm",
analyst = "J. Smith",
reviewer = "A. Jones",
lab = "Analytical Development",
instrument = "Agilent 1260 Infinity II",
# Validation sections (results from earlier in this vignette)
calibration = cal,
lod_loq = lod_loq,
accuracy = accuracy,
precision = list(repeatability = repeatability, intermediate = ip_result),
linearity = linearity,
range = list(lower = 1, upper = 500, units = "ng/mL"),
uncertainty = budget,
# Text sections
specificity = "No interfering peaks observed at the analyte retention time when analyzing blank matrix samples.",
robustness = list(
factors = c("Flow rate (±0.1 mL/min)", "Column temperature (±5°C)", "Mobile phase pH (±0.2)"),
conclusion = "Method showed acceptable robustness within tested parameter ranges."
),
# Conclusions
conclusions = list(
summary = "The analytical method meets all acceptance criteria for precision, accuracy, and linearity.",
recommendations = c(
"Method is suitable for intended use",
"Revalidate if significant changes are made to instrumentation or reagents"
)
),
# References
references = c(
"ICH Q2(R2) Validation of Analytical Procedures (2023)",
"USP <1225> Validation of Compendial Procedures"
)
)
print(report)
#>
#> ── Validation Report ───────────────────────────────────────────────────────────
#> Title: HPLC-UV Method Validation Report
#> Method: Compound X Assay
#> Analyst: J. Smith
#> Lab: Analytical Development
#> Date: 2026-01-01
#>
#> ── Validation Sections ──
#>
#> ℹ Calibration
#> ℹ LOD/LOQ
#> ℹ Accuracy
#> ℹ Precision
#> ℹ Linearity
#> ℹ Range
#> ℹ Specificity/Selectivity
#> ℹ Robustness
#> ℹ Measurement Uncertainty
#>
#> ── Conclusions ──
#>
#> The analytical method meets all acceptance criteria for precision, accuracy,
#> and linearity.
#>
#> ── Provenance ──
#>
#> Generated: 2026-01-01 14:28:29.797234
#> R version: 4.5.2
#> measure version: 0.0.1.9001
#>
#> ℹ Use `render_validation_report()` to generate documentInspecting Report Sections
# Check which sections are included
summary(report)
#>
#> ── Validation Report Summary ───────────────────────────────────────────────────
#> Method: Compound X Assay
#> Date: 2026-01-01
#>
#> # A tibble: 9 × 4
#> section status n_results notes
#> <chr> <chr> <int> <chr>
#> 1 Calibration info 8 ""
#> 2 LOD/LOQ info NA "Method: blank_sd"
#> 3 Accuracy info 3 ""
#> 4 Precision info NA ""
#> 5 Linearity info NA ""
#> 6 Range info NA ""
#> 7 Specificity/Selectivity info NA ""
#> 8 Robustness info NA ""
#> 9 Measurement Uncertainty info NA ""
#>
#> ✔ All sections meet acceptance criteria
# Access specific sections
has_validation_section(report, "calibration")
#> [1] TRUE
has_validation_section(report, "stability") # Not included
#> [1] FALSE
# Get section data
get_validation_section(report, "range")
#> $lower
#> [1] 1
#>
#> $upper
#> [1] 500
#>
#> $units
#> [1] "ng/mL"Adding Custom Sections
You can add additional sections after report creation:
# Add a stability section later
report <- add_validation_section(
report,
"stability",
list(
description = "Short-term stability at room temperature",
results = data.frame(
timepoint = c("0h", "4h", "8h", "24h"),
recovery_pct = c(100, 99.5, 98.8, 97.2)
),
conclusion = "Sample is stable for 24 hours at room temperature."
)
)
has_validation_section(report, "stability")
#> [1] TRUETidy Output
Extract all results as a tidy tibble for further analysis:
tidy(report)
#> # A tibble: 10 × 41
#> section term estimate std_error statistic p_value group n
#> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <chr> <int>
#> 1 Calibration (Int… 0.119 0.0395 3.00 2.39e- 2 NA NA
#> 2 Calibration nomi… 1.06 0.0143 73.8 4.16e-10 NA NA
#> 3 Accuracy NA NA NA NA NA Low 5
#> 4 Accuracy NA NA NA NA NA Mid 5
#> 5 Accuracy NA NA NA NA NA High 5
#> 6 Linearity NA NA NA NA NA NA 15
#> 7 Measurement Uncerta… NA NA NA NA NA NA NA
#> 8 Measurement Uncerta… NA NA NA NA NA NA NA
#> 9 Measurement Uncerta… NA NA NA NA NA NA NA
#> 10 Measurement Uncerta… NA NA NA NA NA NA NA
#> # ℹ 33 more variables: mean_measured <dbl>, mean_reference <dbl>, bias <dbl>,
#> # bias_pct <dbl>, mean_recovery <dbl>, sd_recovery <dbl>, cv_recovery <dbl>,
#> # recovery_ci_lower <dbl>, recovery_ci_upper <dbl>, n_levels <int>,
#> # range_min <dbl>, range_max <dbl>, slope <dbl>, slope_ci_lower <dbl>,
#> # slope_ci_upper <dbl>, intercept <dbl>, intercept_ci_lower <dbl>,
#> # intercept_ci_upper <dbl>, r_squared <dbl>, adj_r_squared <dbl>,
#> # residual_sd <dbl>, residual_cv <dbl>, lof_f <dbl>, lof_p <dbl>, …Rendering Reports
To render a validation report to HTML or PDF, use
render_validation_report(). Two templates are provided:
- ICH Q2(R2): Organized by validation characteristics (specificity, linearity, range, accuracy, precision, etc.)
- USP <1225>: Organized by procedure category (I, II, III, IV)
# Render to HTML using ICH Q2 template (default)
render_validation_report(
report,
output_file = "validation_report.html",
template = "ich_q2"
)
# Render to PDF using USP <1225> template
render_validation_report(
report,
output_file = "validation_report.pdf",
output_format = "pdf",
template = "usp_1225"
)
# Use a custom Quarto template
render_validation_report(
report,
output_file = "custom_report.html",
template_path = "path/to/custom_template.qmd"
)The rendered report includes:
- Header: Method name, date, analyst, reviewer, lab, instrument
- Table of Contents: Auto-generated from sections
- Validation Sections: Each with formatted tables and plots
- Provenance: R version, package versions, timestamp for reproducibility
Summary
The measure package provides a complete toolkit for
analytical method validation:
| Category | Key Functions |
|---|---|
| Calibration |
measure_calibration_fit(),
measure_calibration_predict(),
measure_calibration_verify()
|
| LOD/LOQ |
measure_lod(), measure_loq(),
measure_lod_loq()
|
| Precision |
measure_repeatability(),
measure_intermediate_precision(),
measure_gage_rr()
|
| Accuracy |
measure_accuracy(), measure_linearity(),
measure_carryover()
|
| Method Comparison |
measure_bland_altman(),
measure_deming_regression(),
measure_passing_bablok(),
measure_proficiency_score()
|
| Matrix Effects |
measure_matrix_effect(),
step_measure_standard_addition()
|
| Sample Prep QC |
step_measure_dilution_correct(),
step_measure_surrogate_recovery()
|
| Uncertainty |
measure_uncertainty_budget(),
measure_uncertainty()
|
| Control Charts |
measure_control_limits(),
measure_control_chart()
|
| Criteria |
measure_criteria(), measure_assess(),
criteria_ich_q2(), criteria_bland_altman(),
criteria_matrix_effects()
|
| Drift |
measure_detect_drift(),
step_measure_drift_qc_loess()
|
| Validation Reports |
measure_validation_report(),
render_validation_report()
|
All functions follow a consistent design philosophy: - Tidy
outputs: Results are tibbles with tidy(),
glance(), and autoplot() methods -
Transparent diagnostics: No hidden decisions; all
parameters and flags are visible - Regulatory
compatibility: Designed with ICH, ISO, and FDA guidelines in
mind - Provenance tracking: Audit trails for outlier
handling and data modifications
For more details on any function, see the package documentation with
?function_name.