Creates an Exponentially Modified Gaussian peak model with four parameters: height, center, width (sigma), and tau (exponential decay constant).
Details
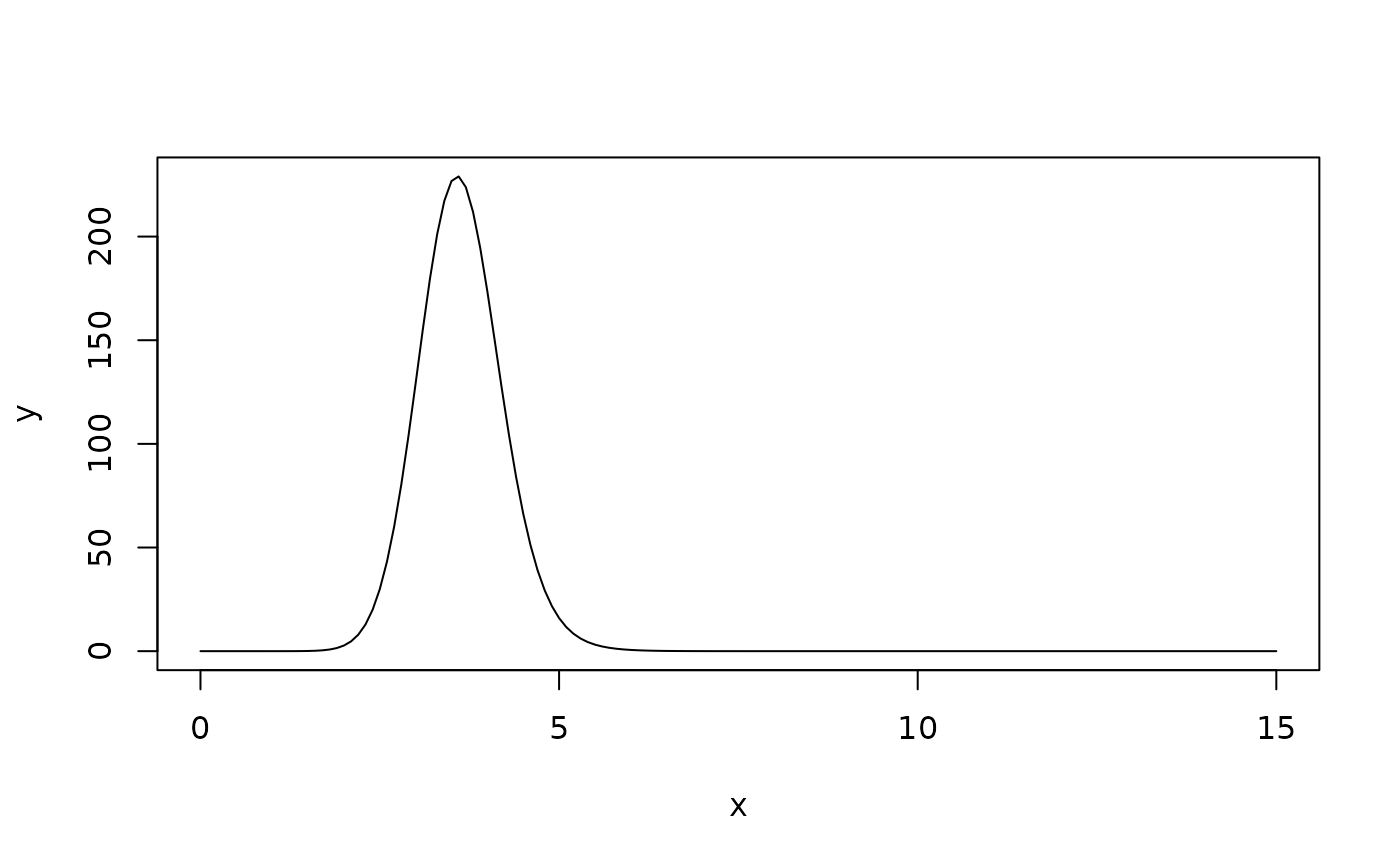
The EMG function models asymmetric peaks with tailing, common in chromatography. It is the convolution of a Gaussian with an exponential decay function.
See also
Other peak-models:
bigaussian_peak_model(),
gaussian_peak_model(),
lorentzian_peak_model()
Examples
model <- emg_peak_model()
x <- seq(0, 15, by = 0.1)
params <- list(height = 1, center = 5, width = 0.5, tau = 0.3)
y <- peak_model_value(model, x, params)
plot(x, y, type = "l")